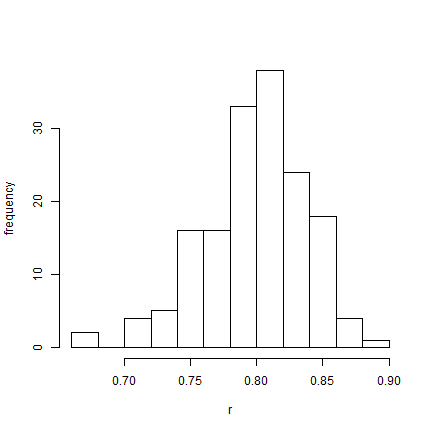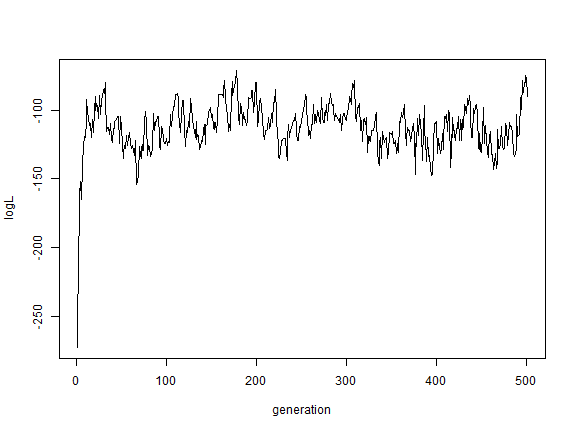Phylogenetic analysis of the threshold model
In this tutorial, we're going to cover two different analyses:
1. We're going to estimate the correlation between two binary
characters, as well as between a binary & a continuous character,
assuming a threshold model of character evolution for our discrete
character; and
2. We're going to reconstruct the ancestral states of a multistate,
naturally ordered character on the tree assuming the threshold model.
If you want to follow along without actually running the simulation and the
MCMC, you can download the following R data object:
threshold.Rdata.
Correlation between discrete & continuous characters under the
threshold model
Here, we're going to use Bayesian MCMC to sample the evolutionary
parameters of our model from their joint posterior probability
distribution. These parameters include evolutionary rates
(σ2s) for each continuous character; and the
correlation between our discrete and continuous character. Since this is
a binary model with a single threshold - we don't have to worry about
the relative positions of the thresholds.
We'll start by simulating a tree & two correlated Brownian variables
on the tree.
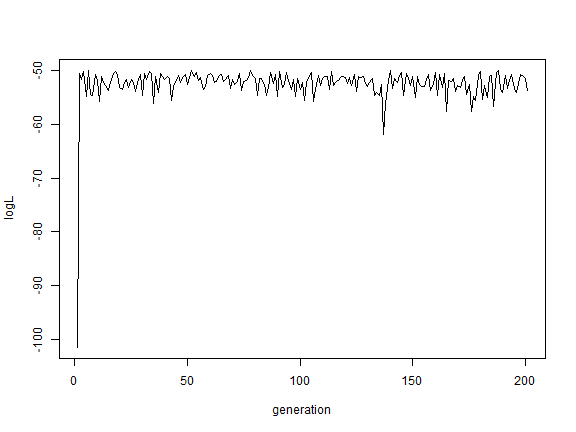
For illustrative purposes, let's start by using threshBayes on two
continuous characters & show that the result lines up closely with our MLE of
the correlation.
library(phytools)
tree <- pbtree(n = 100, scale = 1)
r <- 0.75
V <- matrix(c(1, r, r, 1), 2, 2)
X <- sim.corrs(tree, V)
sample <- 1000
ngen <- 5e+05
burnin <- 0.2 * ngen
AA <- threshBayes(tree, X, types = c("cont", "cont"), ngen = ngen, control = list(sample = sample))
## [1] "gen 1000"
## [1] "gen 2000"
## [1] ...
## [1] "gen 500000"
V.mle <- phyl.vcv(X, vcv(tree), lambda = 1)$R
V.mle[1, 2]/sqrt(V.mle[1, 1] * V.mle[2, 2])
## here is our 'post burnin' mean from the posterior sample
mean(AA$par[(burnin/sample + 1):nrow(AA$par), "r"])
plot(AA$par[, "logL"], type = "l", xlab = "generation", ylab = "logL")
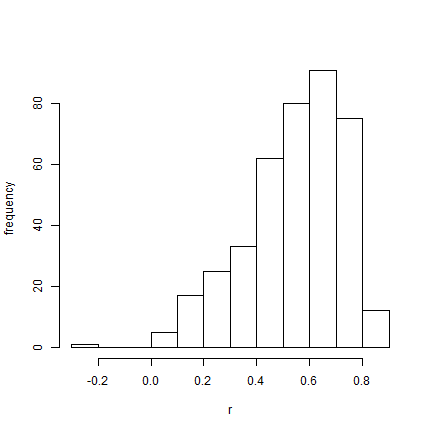
hist(AA$par[(burnin/sample + 1):nrow(AA$par), "r"], xlab = "r", ylab = "frequency",
main = NULL)
OK, this was just a proof-of-concept exercise. Now let's try the same thing
after having converted one of our trait values into a threshold character.
X[, 2] <- as.numeric(sapply(X[, 2], threshState, thresholds = setNames(c(0,
Inf), 0:1)))
BB <- threshBayes(tree, X, types = c("cont", "disc"), ngen = ngen, control = list(sample = sample))
## [1] "gen 1000"
## [1] "gen 2000"
## [1] ...
## [1] "gen 500000"
mean(BB$par[(burnin/sample + 1):nrow(BB$par), "r"])
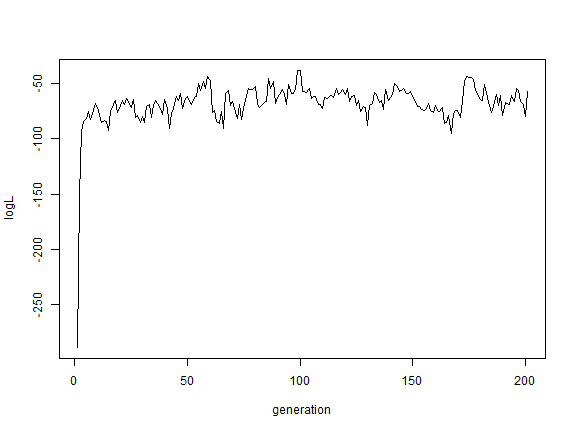
plot(BB$par[, "logL"], type = "l", xlab = "generation", ylab = "logL")
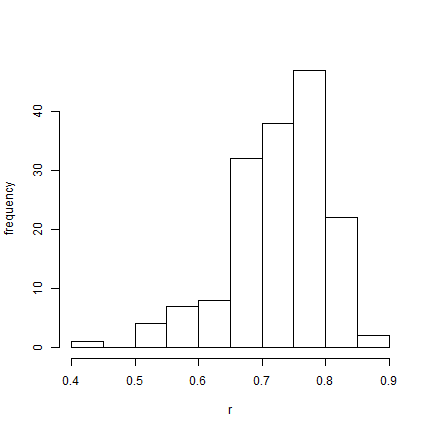
hist(BB$par[(burnin/sample + 1):nrow(BB$par), "r"], xlab = "r", ylab = "frequency",
main = NULL)
Finally, let's analyze two binary characters under the same model. Remember,
each time we're losing information about the underlying correlation between
liabilities. Nonetheless....
X[, 1] <- as.numeric(sapply(X[, 1], threshState, thresholds = setNames(c(0,
Inf), 0:1)))
CC <- threshBayes(tree, X, types = c("disc", "disc"), ngen = ngen, control = list(sample = sample))
## [1] "gen 1000"
## [1] "gen 2000"
## [1] ...
## [1] "gen 500000"
mean(CC$par[(burnin/sample + 1):nrow(CC$par), "r"])
plot(CC$par[, "logL"], type = "l", xlab = "generation", ylab = "logL")
hist(CC$par[(burnin/sample + 1):nrow(CC$par), "r"], xlab = "r", ylab = "frequency",
main = NULL)
So we can see that we can see that threshBayes does a
reasonable job at estimating the correlation between binary & continuous
characters under the threshold model.
## Loading required package: coda
rA <- AA$par[(burnin/sample + 1):nrow(AA$par), "r"]
class(rA) <- "mcmc"
effectiveSize(rA)
## lower upper
## var1 0.663 0.8319
## attr(,"Probability")
## [1] 0.9501
rB <- BB$par[(burnin/sample + 1):nrow(BB$par), "r"]
class(rB) <- "mcmc"
effectiveSize(rB)
## lower upper
## var1 0.3522 0.8179
## attr(,"Probability")
## [1] 0.9501
rC <- CC$par[(burnin/sample + 1):nrow(CC$par), "r"]
class(rC) <- "mcmc"
effectiveSize(rC)
## lower upper
## var1 0.1622 0.8076
## attr(,"Probability")
## [1] 0.9501
Ancestral character estimation under the threshold model
We can also do ancestral character reconstruction for discrete characters
using the threshold model.
n <- 100
ngen <- 1e+06
sample <- 1000
tree <- pbtree(n = n, scale = 10)
x <- fastBM(tree, sig2 = 1, a = 0.5, internal = TRUE)
th <- sapply(x, threshState, thresholds = setNames(c(0, 0.5, 2, Inf), letters[1:4]))
mcmc <- ancThresh(tree, th[1:n], ngen = ngen, sequence = letters[1:4], control = list(sample = sample,
plot = FALSE))
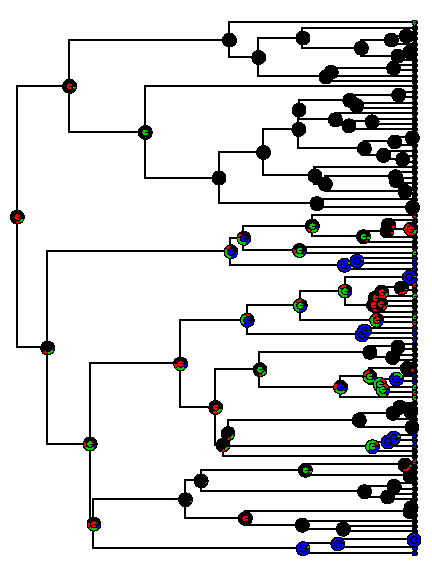
plotTree(tree, setEnv = TRUE, ftype = "off")
## setEnv=TRUE is experimental. please be patient with bugs
tiplabels(pie = to.matrix(th[1:n], letters[1:4]), piecol = palette()[1:4], cex = 0.3)
nodelabels(pie = mcmc$ace, piecol = palette()[1:4], cex = 0.8)
nodelabels(pie = to.matrix(th[1:tree$Nnode + n], letters[1:4]), piecol = palette()[1:4],
cex = 0.4)
Theoretically, we can even use this method to identify the relative positions of
thresholds:
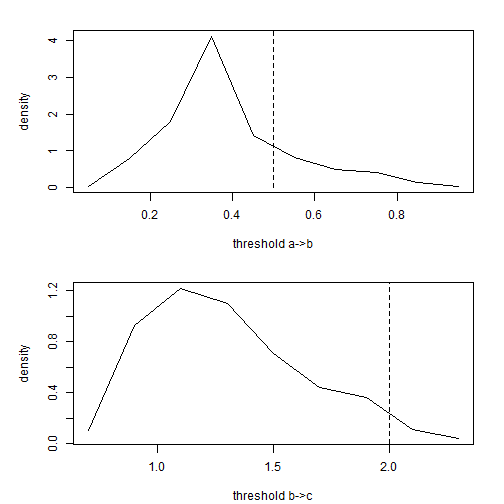
colMeans(mcmc$par[(0.2 * ngen/sample):(ngen/sample) + 1, letters[1:4]])
## a b c d
## 0.0000 0.3878 1.2953 Inf
par(mfrow = c(2, 1))
par(mar = c(4.1, 5.1, 2.1, 2.1))
xx <- hist(mcmc$par[(0.2 * ngen/sample):(ngen/sample) + 1, letters[2]], plot = FALSE)
plot(xx$mids, xx$density, type = "l", xlab = "threshold a->b", ylab = "density")
lines(c(0.5, 0.5), c(0, 1.1 * max(xx$density)), lty = "dashed")
xx <- hist(mcmc$par[(0.2 * ngen/sample):(ngen/sample) + 1, letters[3]], plot = FALSE)
plot(xx$mids, xx$density, type = "l", xlab = "threshold b->c", ylab = "density")
lines(c(2, 2), c(0, 1.1 * max(xx$density)), lty = "dashed")
That's it!
Written by Liam J. Revell. Last updated Aug. 10, 2013