## first load phytools require(phytools) ## simulate a tree tree <- pbtree(n = 26, tip.label = LETTERS) plotTree(tree)
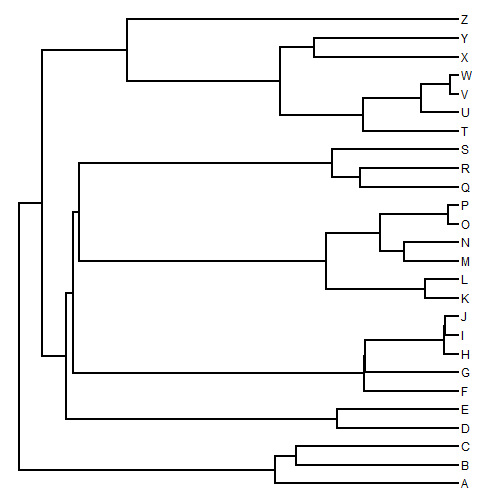
In this tutorial I'm going to quickly overview a range of plotting methods for phylogenies & comparative data that are implemented in the phytools package.
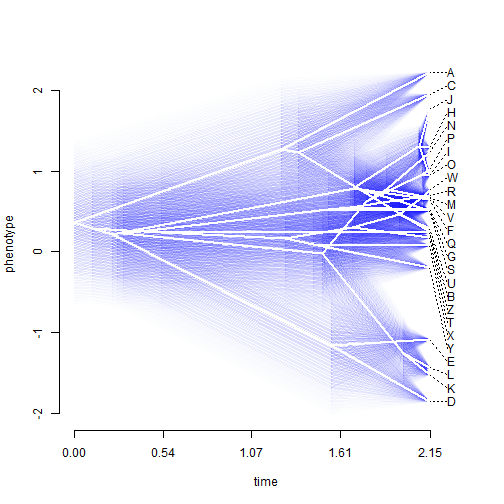
The first plotting method I'll illustrate is called a 'traitgram' which is a projection of the tree into a space defined by trait values & time since the root. So, for example:
## first load phytools require(phytools) ## simulate a tree tree <- pbtree(n = 26, tip.label = LETTERS) plotTree(tree)

## simulate data under Brownian motion x <- fastBM(tree) x
## A B C D E F G H ## 2.21950 0.25627 1.94721 -1.85028 -1.08259 0.64343 0.51627 1.29706 ## I J K L M N O P ## 0.95156 1.76106 -1.51259 -1.43161 0.67923 1.23026 0.94695 1.02401 ## Q R S T U V W X ## 0.52726 0.70021 0.51362 0.17891 0.28827 0.66855 0.75961 0.07452 ## Y Z ## -0.19691 0.24588
## plot a simple traitgram phenogram(tree, x)
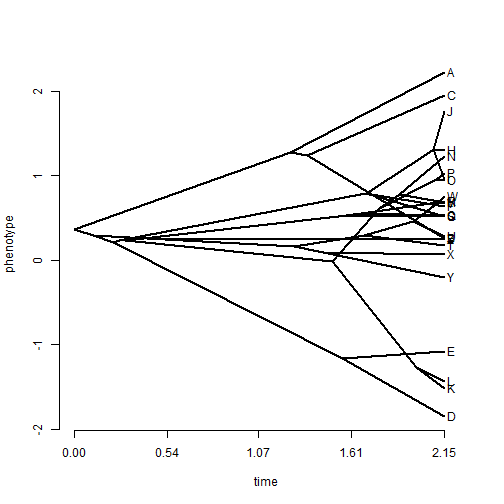
## tip labels are messy - spread them phenogram(tree, x, spread.labels = TRUE)
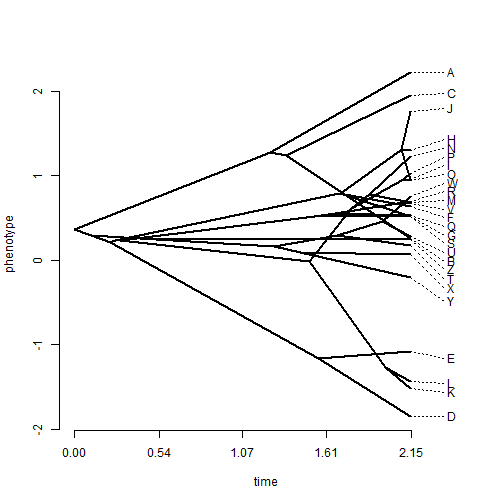
## change the spread costs phenogram(tree, x, spread.labels = TRUE, spread.cost = c(1, 0))
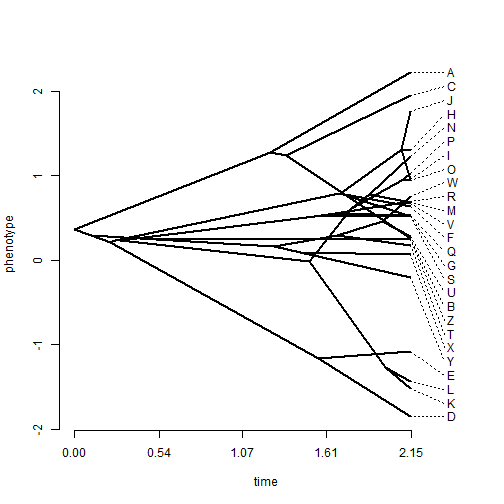
We can also plot a traitgram with the uncertainty about ancestral character states visualized using transparent probability density. This is implemented in the phytools function fancyTree. So, for instance:
## plot traitgram with 95% CI fancyTree(tree, type = "phenogram95", x = x, spread.cost = c(1, 0))
Next, we can explore the continuous character mapping function in phytools called contMap. Here is a quick demo using the same data:
## plot contMap obj <- contMap(tree, x)
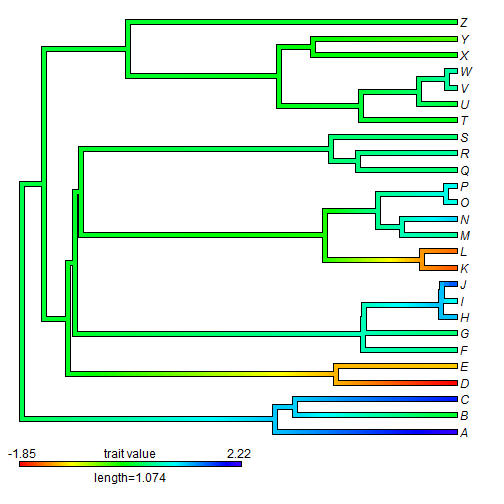
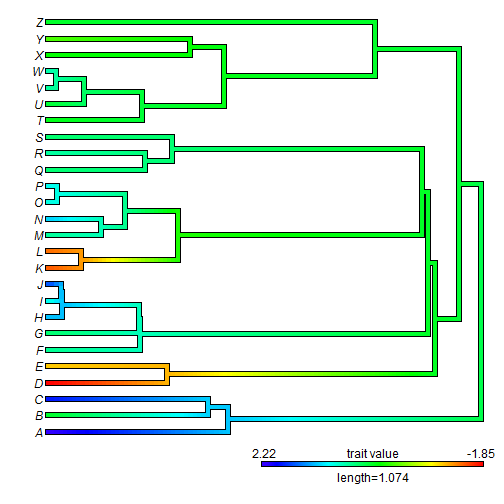
contMap returns an object of class "contMap" which we can then more easily replot using, for instance, different parameters:
## plot leftward plot(obj, direction = "leftwards")
## plot fan style tree plot(obj, type = "fan", lwd = 5) ## for example
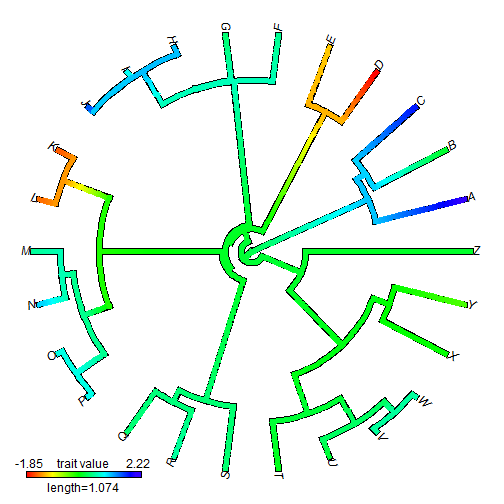
Here are some real data using anole.tre and svl.csv:
anole.tree <- read.tree("anole.tre") ## or ## anole.tree<-read.tree('http://www.phytools.org/anthrotree/plot/anole.tre') anole.tree
## ## Phylogenetic tree with 100 tips and 99 internal nodes. ## ## Tip labels: ## ahli, allogus, rubribarbus, imias, sagrei, bremeri, ... ## ## Rooted; includes branch lengths.
svl <- read.csv("svl.csv", header = TRUE, row.names = 1) ## of ## svl<-read.csv('http://www.phytools.org/anthrotree/plot/svl.csv',header=TRUE,row.names=1) svl <- as.matrix(svl)[, 1] svl
## ahli alayoni alfaroi aliniger ## 4.039 3.816 3.527 4.037 ## allisoni allogus altitudinalis alumina ## 4.375 4.040 3.843 3.589 ## alutaceus angusticeps argenteolus argillaceus ## 3.555 3.789 3.971 3.758 ## armouri bahorucoensis baleatus baracoae ## 4.122 3.827 5.053 5.043 ## barahonae barbatus barbouri bartschi ## 5.077 5.004 3.664 4.281 ## bremeri breslini brevirostris caudalis ## 4.113 4.051 3.874 3.912 ## centralis chamaeleonides chlorocyanus christophei ## 3.698 5.042 4.275 3.885 ## clivicola coelestinus confusus cooki ## 3.759 4.298 3.938 4.092 ## cristatellus cupeyalensis cuvieri cyanopleurus ## 4.190 3.462 4.875 3.630 ## cybotes darlingtoni distichus dolichocephalus ## 4.211 4.302 3.929 3.909 ## equestris etheridgei eugenegrahami evermanni ## 5.114 3.658 4.129 4.166 ## fowleri garmani grahami guafe ## 4.289 4.769 4.154 3.877 ## guamuhaya guazuma gundlachi haetianus ## 5.037 3.764 4.188 4.317 ## hendersoni homolechis imias inexpectatus ## 3.860 4.033 4.100 3.537 ## insolitus isolepis jubar krugi ## 3.800 3.657 3.953 3.887 ## lineatopus longitibialis loysiana lucius ## 4.129 4.242 3.701 4.199 ## luteogularis macilentus marcanoi marron ## 5.101 3.716 4.079 3.832 ## mestrei monticola noblei occultus ## 3.987 3.771 5.083 3.663 ## olssoni opalinus ophiolepis oporinus ## 3.794 3.838 3.638 3.846 ## paternus placidus poncensis porcatus ## 3.803 3.774 3.820 4.259 ## porcus pulchellus pumilis quadriocellifer ## 5.038 3.799 3.467 3.902 ## reconditus ricordii rubribarbus sagrei ## 4.483 5.014 4.078 4.067 ## semilineatus sheplani shrevei singularis ## 3.697 3.683 3.983 4.058 ## smallwoodi strahmi stratulus valencienni ## 5.035 4.274 3.870 4.322 ## vanidicus vermiculatus websteri whitemani ## 3.626 4.803 3.917 4.097
obj <- contMap(anole.tree, svl, fsize = c(0.6, 1), outline = FALSE)
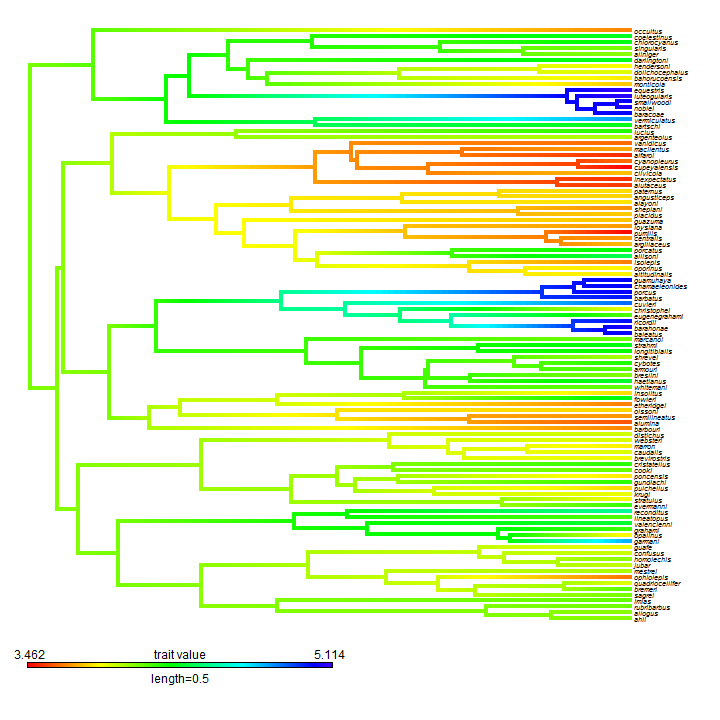
We can also plot this using a circular or fan tree:
plot(obj, type = "fan", outline = FALSE)
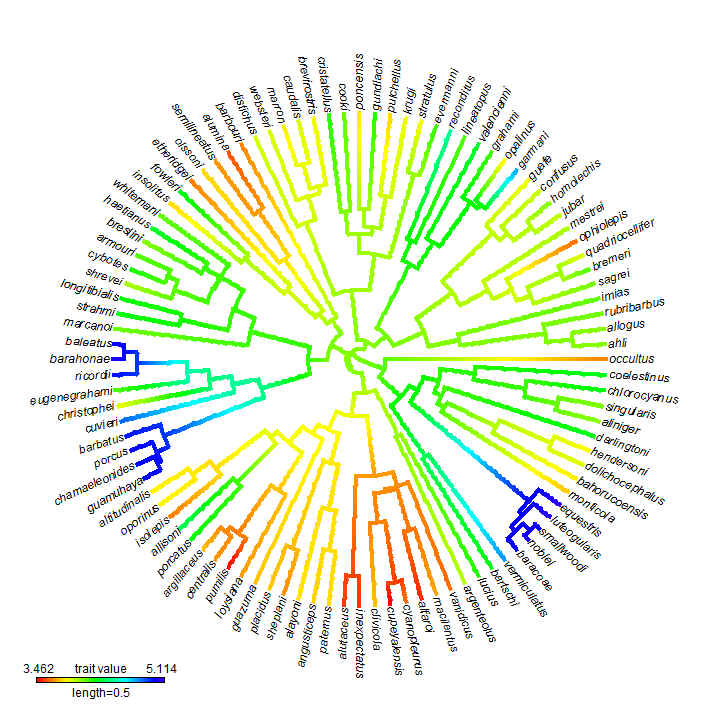
Note that the intense aliasing is just a byproduct of plotting to .png format. To get a high quality image we can plot to .pdf or other formats. For instance:
pdf(file = "anole.contMap.pdf", width = 10, height = 10) plot(obj, type = "fan", outline = FALSE) dev.off()
## pdf ## 2
The result can be viewed here.
Finally, a relatively simple new plotting method in phytools is the function plotTree.wBars. This function is not in the current CRAN release of phytools, so to run it you will need to first download & install phytools from source. That function pretty much does what it sounds like it does:
plotTree.wBars(anole.tree, exp(svl), type = "fan", scale = 0.002)
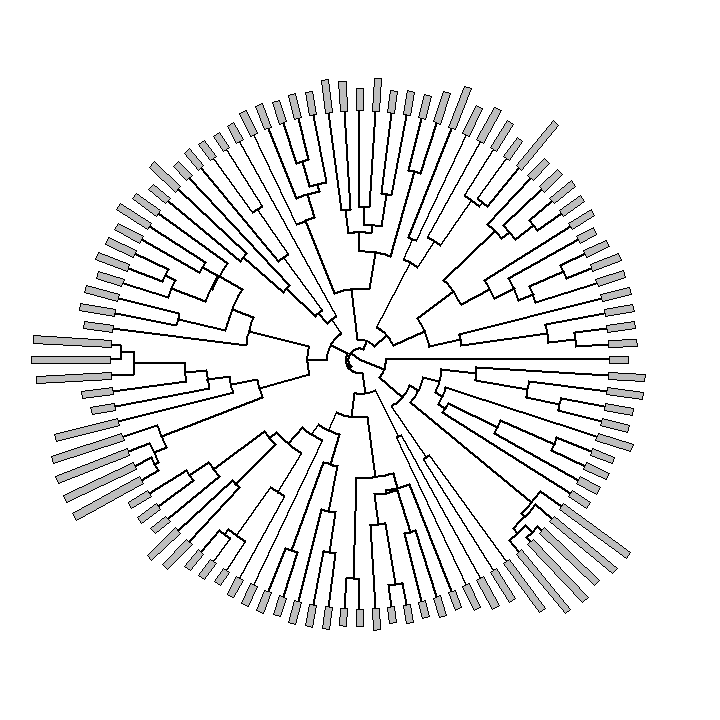
This is also kind of cool:
obj <- contMap(anole.tree, exp(svl), plot = FALSE) plotTree.wBars(obj$tree, exp(svl), method = "plotSimmap", colors = obj$cols, type = "fan", scale = 0.002) add.color.bar(1, obj$cols, title = "trait value", lims = obj$lims, prompt = FALSE, x = 0.9 * par()$usr[1], y = 0.9 * par()$usr[3])
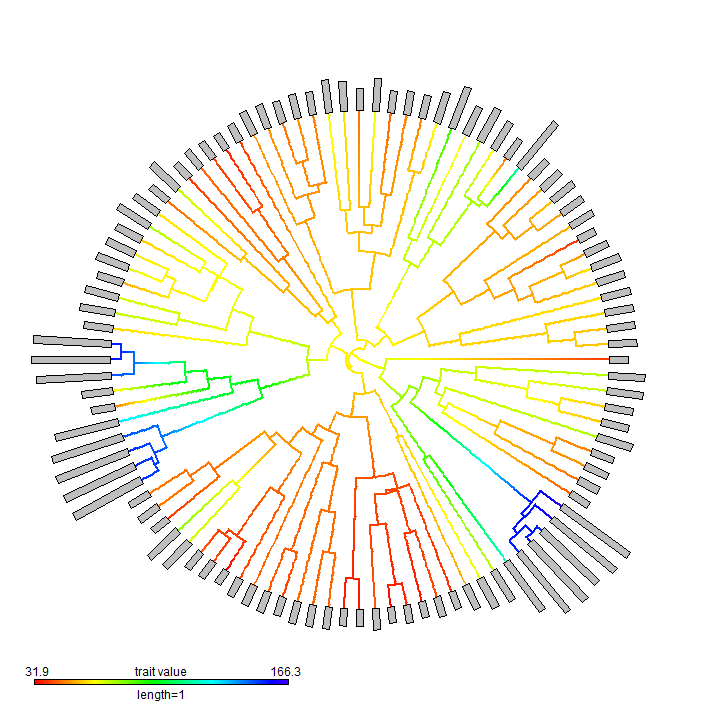
pdf(file = "anole.plotTree.wBars.pdf", width = 10, height = 10) plotTree.wBars(obj$tree, exp(svl), method = "plotSimmap", colors = obj$cols, type = "fan", scale = 0.002) add.color.bar(1, obj$cols, title = "trait value", lims = obj$lims, prompt = FALSE, x = 0.9 * par()$usr[1], y = 0.9 * par()$usr[3]) dev.off()
## pdf ## 2
(A non-aliased version here.)
We can also do two dimensional visualizations of phylogenies in morphospace. The is called a 'phylomorphospace'. E.g.:
X <- fastBM(tree, nsim = 3) ## simulate 3 characters phylomorphospace(tree, X[, c(1, 2)], xlab = "trait 1", ylab = "trait 2")
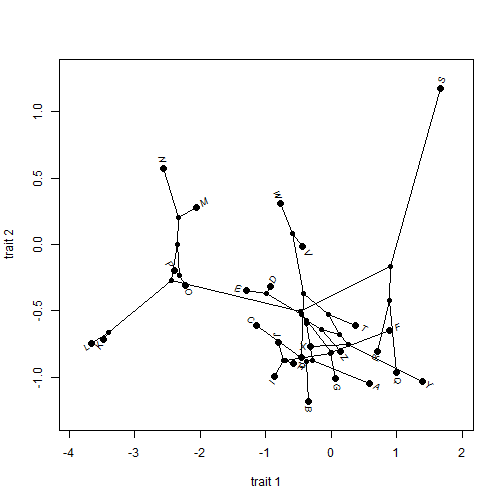
With phytools we can do static or animated version of the same. I'll just do a static version here, but animated is the default:
phylomorphospace3d(tree, X, method = "static")
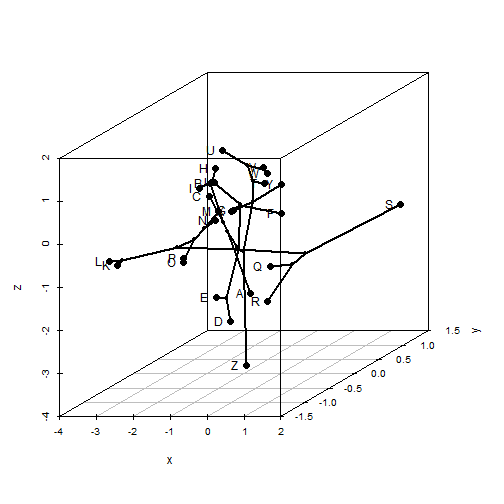
Try the animated version on your own.
Finally, it's possible to combine the continuous character mapping and 2D phylomorphospaces using a type of phylogenetic scatterplot. Here's a demo:
fancyTree(tree, type = "scattergram", X = X)
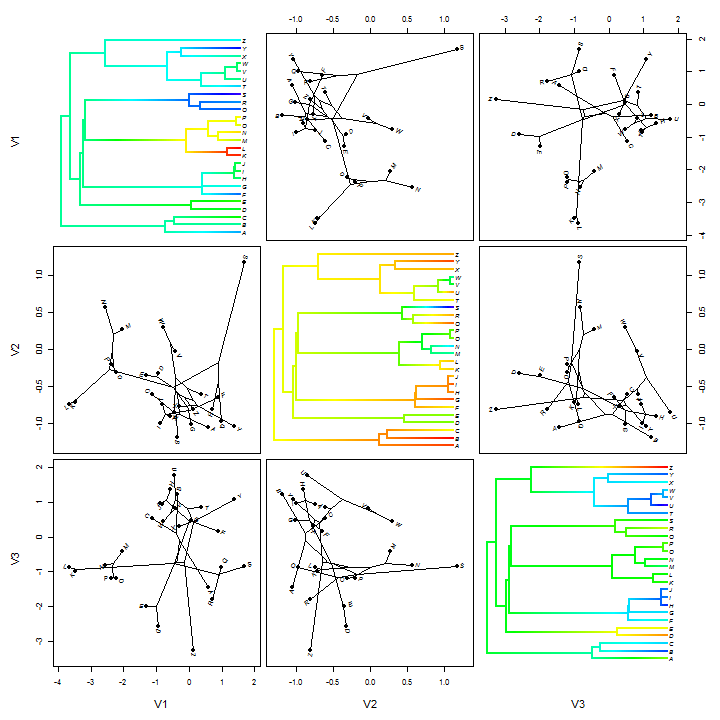
Stochastic character maps are easy to plot using phytools. For instance:
data(anoletree) plotSimmap(anoletree, type = "fan")
## no colors provided. using the following legend: ## CG GB TC TG Tr Tw ## "black" "red" "green3" "blue" "cyan" "magenta"
## setEnv=TRUE for this type is experimental. please be patient with bugs
## we can use any color scheme, but this is the default cols <- setNames(palette()[1:length(unique(getStates(anoletree, "tips")))], sort(unique(getStates(anoletree, "tips")))) add.simmap.legend(colors = cols, x = 0.9 * par()$usr[1], y = 0.9 * par()$usr[4], prompt = FALSE)
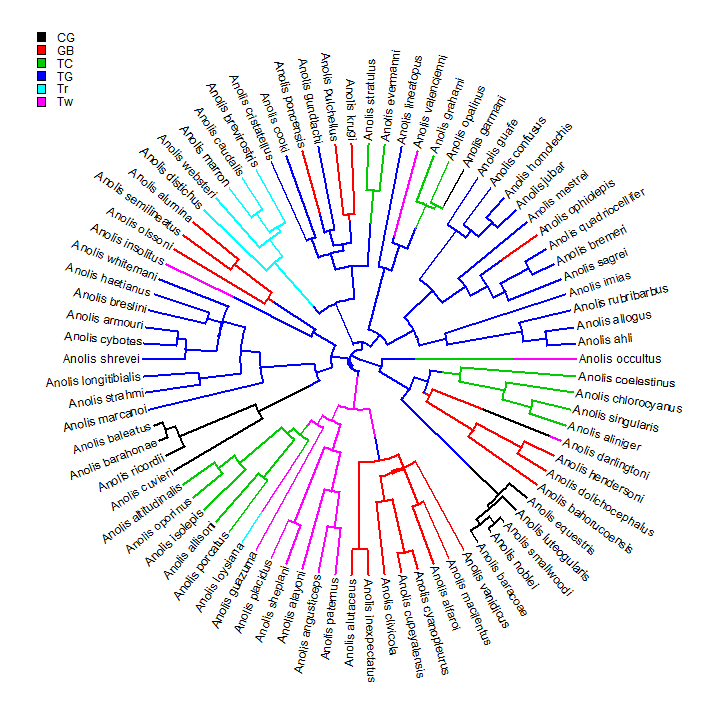
This is just one stochastically mapped tree. Let's perform the method of stochastic mapping & plot the posterior probabilities at internal nodes.
x <- getStates(anoletree, "tips") trees <- make.simmap(anoletree, x, model = "ER", nsim = 100)
## CG GB TC TG Tr Tw ## CG -0.11571 0.02314 0.02314 0.02314 0.02314 0.02314 ## GB 0.02314 -0.11571 0.02314 0.02314 0.02314 0.02314 ## TC 0.02314 0.02314 -0.11571 0.02314 0.02314 0.02314 ## TG 0.02314 0.02314 0.02314 -0.11571 0.02314 0.02314 ## Tr 0.02314 0.02314 0.02314 0.02314 -0.11571 0.02314 ## Tw 0.02314 0.02314 0.02314 0.02314 0.02314 -0.11571
## CG GB TC TG Tr Tw ## 0.1667 0.1667 0.1667 0.1667 0.1667 0.1667
obj <- describe.simmap(trees, plot = FALSE) obj
## 100 trees with a mapped discrete character with states: ## CG, GB, TC, TG, Tr, Tw ## ## trees have 25.59 changes between states on average ## ## changes are of the following types: ## CG,GB CG,TC CG,TG CG,Tr CG,Tw GB,CG GB,TC GB,TG GB,Tr GB,Tw TC,CG ## x->y 0.25 0.31 0.25 0.2 0.32 0.71 1.02 1.2 0.51 1.46 1.25 ## TC,GB TC,TG TC,Tr TC,Tw TG,CG TG,GB TG,TC TG,Tr TG,Tw Tr,CG Tr,GB ## x->y 0.67 0.36 0.69 1.01 1.13 3.25 1.98 1.11 1.66 0.18 0.34 ## Tr,TC Tr,TG Tr,Tw Tw,CG Tw,GB Tw,TC Tw,TG Tw,Tr ## x->y 0.23 0.22 0.45 0.65 1.26 1.49 0.8 0.63 ## ## mean total time spent in each state is: ## CG GB TC TG Tr Tw total ## raw 13.09759 46.1139 32.5794 69.7658 12.91992 31.1904 205.7 ## prop 0.06368 0.2242 0.1584 0.3392 0.06282 0.1517 1.0
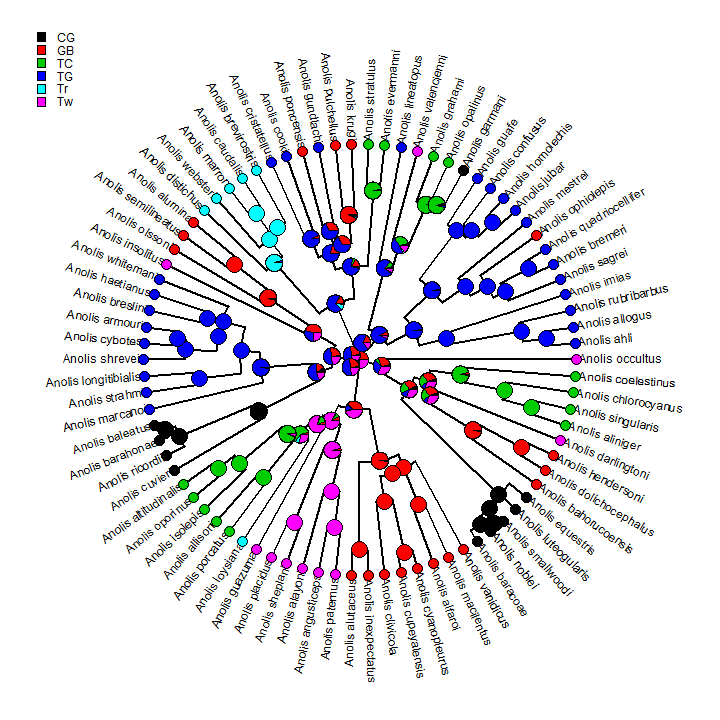
plot(obj, type = "fan")
## setEnv=TRUE for this type is experimental. please be patient with bugs
add.simmap.legend(colors = cols, x = 0.9 * par()$usr[1], y = 0.9 * par()$usr[4], prompt = FALSE)
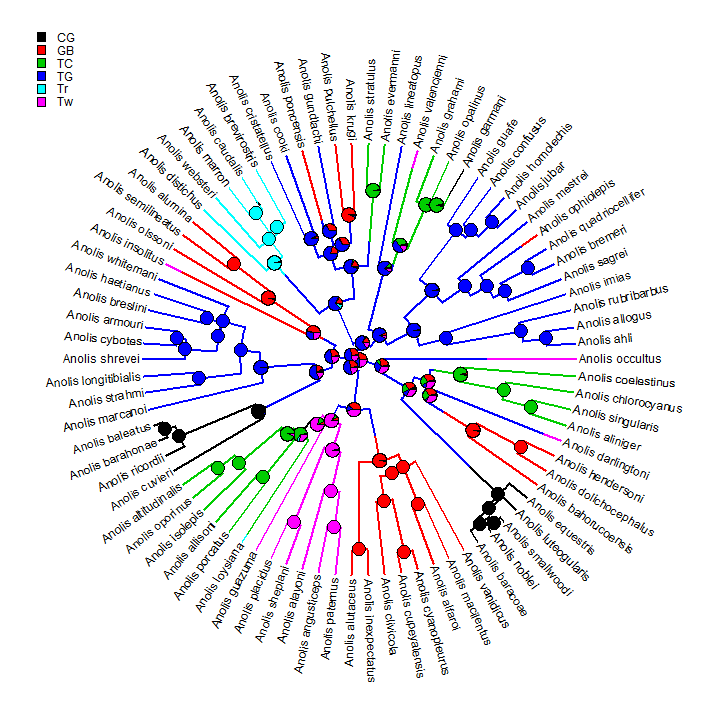
We can also overlay the posterior probabilities on a random stochastic map in a similar way:
plotSimmap(trees[[1]], type = "fan")
## no colors provided. using the following legend: ## CG GB TC TG Tr Tw ## "black" "red" "green3" "blue" "cyan" "magenta"
## setEnv=TRUE for this type is experimental. please be patient with bugs
nodelabels(pie = obj$ace, piecol = cols, cex = 0.5) add.simmap.legend(colors = cols, x = 0.9 * par()$usr[1], y = 0.9 * par()$usr[4], prompt = FALSE)
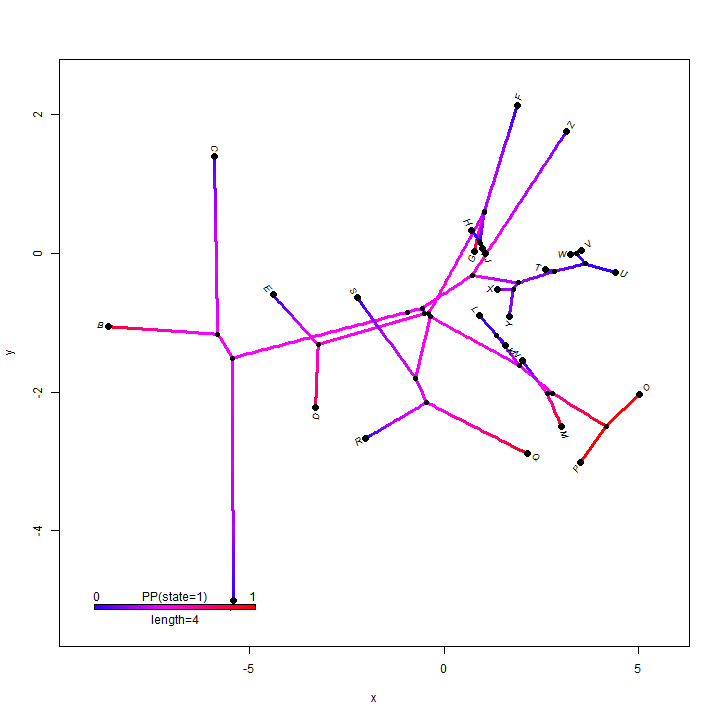
For binary characters, we can also map the posterior density of stochastic maps onto the nodes & branches of a tree using the phytools function densityMap. Here's a demo of this using simulated data.
## first let's simulate data Q <- matrix(c(-1, 1, 1, -1), 2, 2) rownames(Q) <- colnames(Q) <- c(0, 1) tree <- sim.history(tree, Q) x <- tree$states x
## A B C D E F G H I J K L M N O P Q R ## "0" "1" "0" "1" "0" "0" "1" "0" "0" "0" "0" "0" "1" "0" "1" "1" "1" "0" ## S T U V W X Y Z ## "0" "0" "0" "0" "0" "0" "0" "0"
## stochastic maps trees <- make.simmap(tree, x, nsim = 100)
## 0 1 ## 0 -1.201 1.201 ## 1 1.201 -1.201
## 0 1 ## 0.5 0.5
## make densityMap obj <- densityMap(trees, lwd = 4, outline = TRUE)
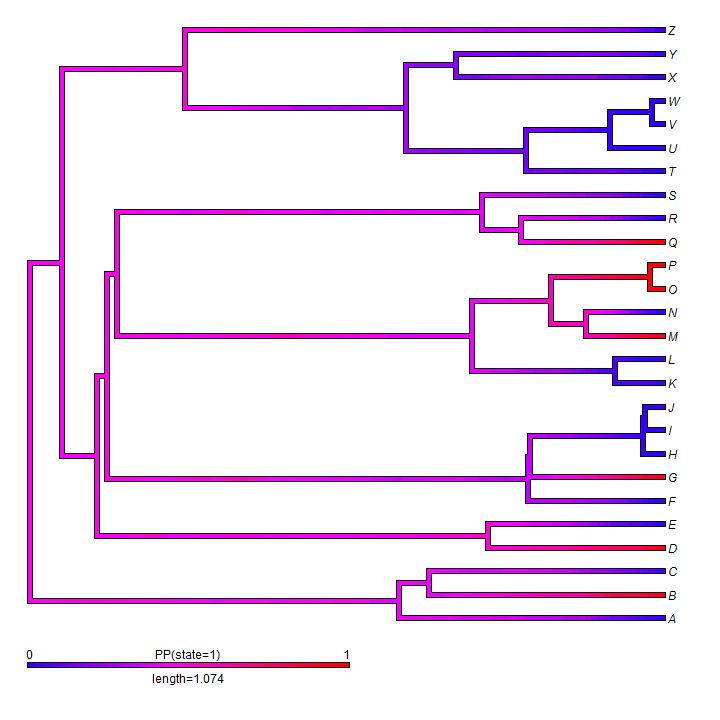
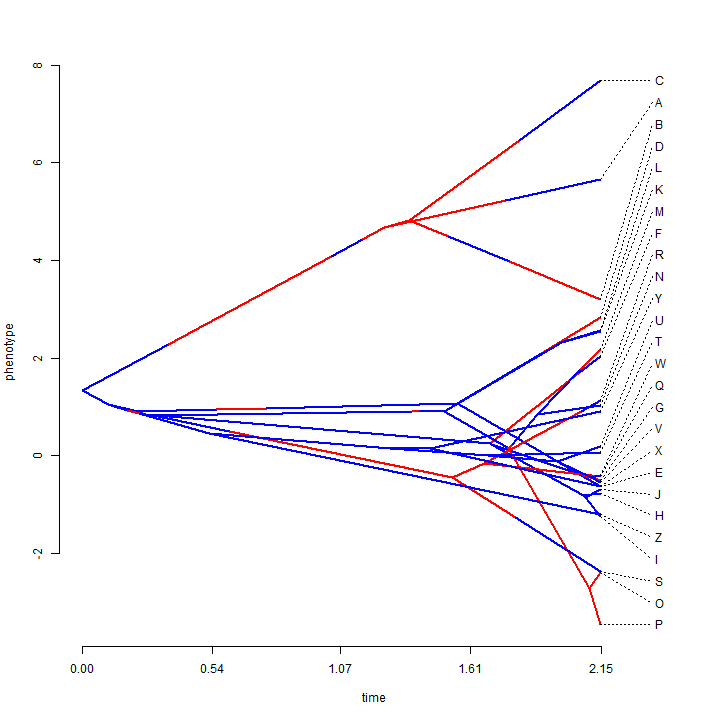
We can, for instance, overlay a discrete character history (or stochastic map) onto a traitgram or phylomorphospace.
## simulate data with a high rate on some branches x <- sim.rates(tree, setNames(c(1, 10), c(0, 1))) ## plot traitgram phenogram(tree, x, colors = setNames(c("blue", "red"), c(0, 1)), spread.labels = TRUE, spread.cost = c(1, 0))
## we can do the same with phylomorphospaces X <- cbind(sim.rates(tree, setNames(c(1, 10), c(0, 1))), sim.rates(tree, setNames(c(1, 10), c(0, 1)))) phylomorphospace(tree, X, colors = setNames(c("blue", "red"), c(0, 1)))
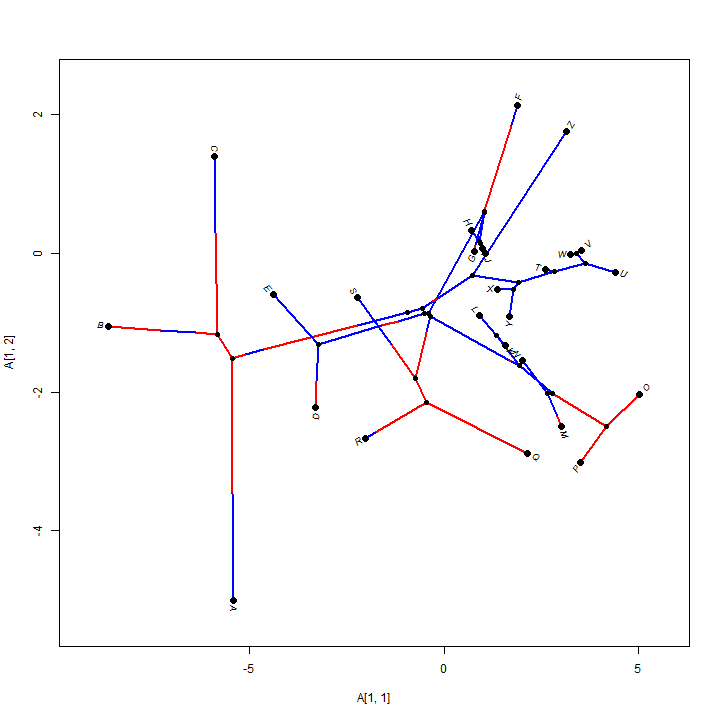
## it is even possible to overlay a posterior density from stochastic mapping phylomorphospace(obj$tree, X, colors = obj$cols, lwd = 3, xlab = "x", ylab = "y") add.color.bar(4, obj$cols, title = "PP(state=1)", prompt = FALSE, x = 0.9 * par()$usr[1], y = 0.9 * par()$usr[3])
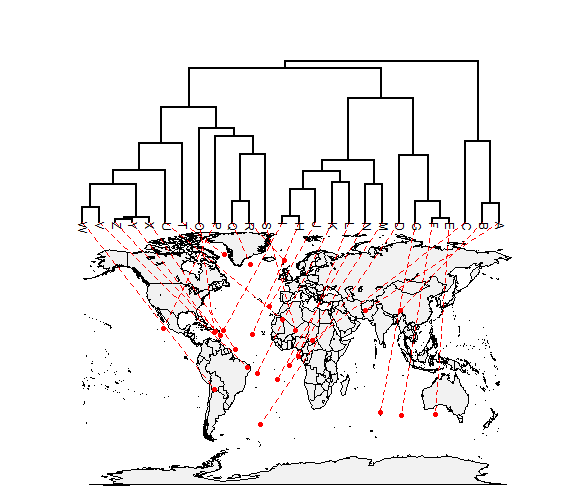
How about projecting a tree onto a geographic map? Here's an example using phytools function phylo.to.map:
# simulate tree & data tree <- pbtree(n = 26, scale = 100) tree$tip.label <- LETTERS[26:1] lat <- fastBM(tree, sig2 = 10, bounds = c(-90, 90)) long <- fastBM(tree, sig2 = 80, bounds = c(-180, 180)) # now plot projection xx <- phylo.to.map(tree, cbind(lat, long), plot = FALSE)
## tree points to map coordinates plot(xx, type = "phylogram", asp = 1.3, mar = c(0.1, 0.5, 3.1, 0.1))
## tree projected directly on map plot(xx, type = "direct", asp = 1.3, mar = c(0.1, 0.5, 3.1, 0.1))
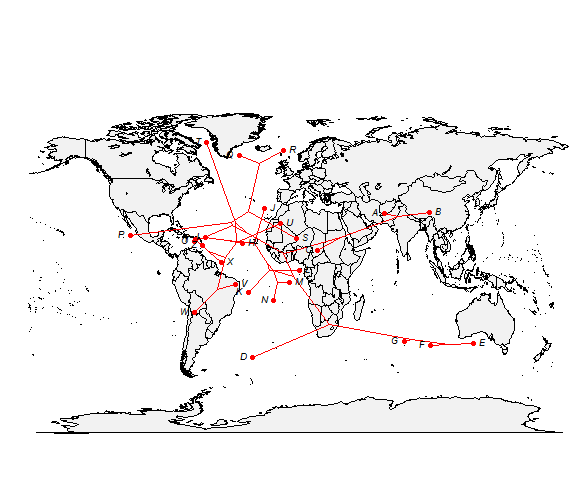
That's all for now!
Written by Liam J. Revell. Last updated May 29, 2014